2. 上海海洋大学水产与生命学院, 上海 201306
2. College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306, China
细菌群落是河流生态系统的重要组成部分, 在河流污染物降解和转化中起着关键作用[1]。已有研究表明, 浮游细菌会对生境中生物因子和非生物因子的变化做出响应[2]。因此, 细菌群落结构的动态变化被认为是衡量水生态环境的重要指标之一[3]。几十年来, 微生物生态学家一直面临的挑战是如何根据环境DNA扩增文库中有代表性的中等大小rRNA数据集, 推断微生物群落的组成[4]。随着分子生物学的发展, 非培养的研究方法成为微生物研究的主要方法, 例如利用变性梯度凝胶电泳(denatured gradient gel electrophoresis, DGGE)、稳定性同位素探测、基因芯片和高通量测序等方法研究人体、土壤、海洋的微生物多样性等取得大量成果[5]。高通量测序技术以微生物目标基因的PCR产物为样本进行测序, 不仅能从群落水平上解释微生物的组成变化, 还能从分类学上显示群落的具体变化, 在环境微生物研究中应用广泛[6-7]。然而目前水体微生物的研究主要集中于海洋和湖泊[8-9], 对于河流的研究相对较少。
珠江河网主要由西江、北江、东江的干流、支流汇聚而成, 河道密布, 流经广州及周边城市的居民区及工农业生产区。由于受季风气候的影响, 珠江流域降水量约80%集中在丰水期, 水资源时空分布极不均匀[10]。珠江河网流域面积仅占珠江水系的5.9%, 但废污水排放量约占总排污量的50%[11], 根据对珠江河网区域的监测发现, 该区域许多水体总氮含量长期处于劣Ⅴ类水平, 总磷含量一直处于Ⅱ~Ⅲ类水平[12]。相对于春夏秋季, 冬季的降水量最少, 径流量等自然因素对珠江河网的干扰作用降到最低, 而人为作用对环境的影响则较为突出。水质的改变不仅影响浮游生物、鱼类和底栖生物的生存状态和种群资源量, 也会对浮游细菌的群落结构产生影响。
基于水体生态修复中微生物群落的重要地位以及高通量测序分析微生物群落结构的快速发展, 本研究以珠江河网为研究区域, 利用高通量测序分析浮游细菌的群落结构组成。利用冗余分析方法(redundancy analysis, RDA)对其与环境因子的关系进行分析, 进行Pearson相关分析确定细菌多样性及优势群落与环境因子的关系, 探讨决定细菌群落的可能因素。
1 材料与方法 1.1 采样地点及样品采集调查区域为珠三角河网水域(112.18°E~ 113.51°E, 22.38°N~23.17°N), 一共布设13个位点(图 1), 包括北滘(BJ)、陈村(CC)、横沥(HL)、榄核(LH)、莲花山(LHS)、青岐(QQ)、市桥(SQ)、外海(WH)、小榄(XL)、小塘(XT)、新围(XW)、珠江桥(ZJQ)、左滩(ZT)。
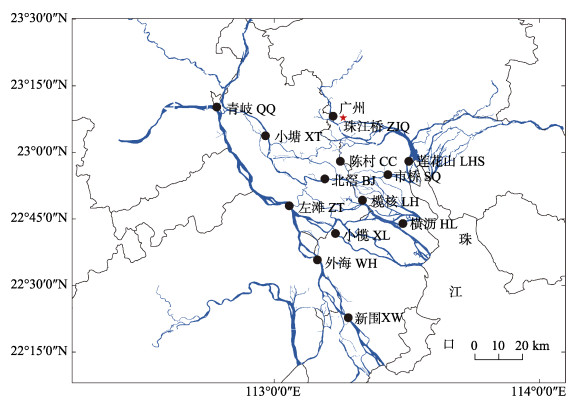
|
图 1 珠江河网调查区域示意图 北滘(113.19°E, 22.90°N), 陈村(113.25°E, 22.97°N), 横沥(113.48°E, 22.73°N), 榄核(113.33°E, 22.82°N), 莲花山(113.51°E, 22.97°N), 青岐(112.18°E, 23.17°N), 市桥(113.43°E, 22.92°N), 外海(113.16°E, 22.59°N), 小榄(113.41°E, 22.44°N), 小塘(112.97°E, 23.06°N), 新围(113.27°E, 22.38°N), 珠江桥(22.97°N, 23.14°N), 左滩(113.06°E, 22.80°N). Fig.1 Map of the Pearl River Delta BJ-Beijiao, 113.19°E, 22.90°N; CC-Chencun, 113.25°E, 22.97°N; HL-Hengli, 113.48°E, 22.73°N; LH-Lanhe, 113.33°E, 22.82°N; LHS-Lianhuashan, 113.51°E, 22.97°N; QQ-Qingqi, 112.18°E, 23.17°N; SQ-Shiqiao, 113.43°E, 22.92°N; WH-Waihai, 113.16°E, 22.59°N; XL-Xiaolan, 113.41°E, 22.44°N; XT-Xiaotang, 112.97°E, 23.06°N; XW-Xinwei, 113.27°E, 22.38°N; ZJQ-Zhujiangqiao, 22.97°N, 23.14°N; ZT-Zuotan, 113.06°E, 22.80°N. |
实验样品为2017年11月及2018年1月两次采集的表层水样(水深0.5 m), 使用5 L的有机玻璃采水器采集表层水样3 L。其中2 L样本采集后立即转移到无菌取样袋中(TWIRLʹEM®, LABPLAS INC., Quebec, Canada), 在4℃冷藏条件下尽快运回实验室, 经过2 µm聚砜超滤膜和0.22 µm滤膜(Pall, USA)过滤后, 将滤膜迅速转移至10 mL离心管, 进行DNA提取之前一直在−20℃保存。其余样品等量分成两份:一份经0.45 µm孔径醋酸纤维素滤膜(Whatman, USA)过滤后保存在样品瓶中, 另一份加入浓硫酸进行固定; 分别在4℃条件下储存用于化学测定。
1.2 物理化学测定环境参数包括温度(temperature, T, ℃)、pH、盐度(salinity, S)、溶解氧(dissolved oxygen, DO, mg/L)、总溶解性固体(total dissolved solid, TDS, mg/L)和氧化还原电位(oxidation-reduction potential, ORP, mV), 采用YSI多参数水质分析仪(YSI Inc., USA)进行现场监测。采用经0.45 µm孔径醋酸纤维素滤膜(Whatman, USA)过滤的样品, 用SAN++全自动水化学分析仪(Skalar Analytical B.V., the Netherlands)分别在540 nm、540 nm、660 nm、880 nm和810 nm波长下测定地表水中硝酸盐(NO3‒)、亚硝酸盐(NO2‒)、氨(NH4+)、磷酸盐(PO43‒)和硅酸盐(SiO32-的浓度, 加浓硫酸样品分别在540 nm、880 nm波长下测定总氮(TN)、总磷(TP)的浓度。
1.3 DNA提取、PCR扩增及上机测序使用DNA提取试剂盒(Magen Hipure Spil DNA Kit)从样本中提取DNA, 使用Qubit® dsDNA HS Assay Kit (Qubit® dsDNA HS Assay Kit)检测DNA浓度。DNA提取与检测流程参照两种试剂盒的使用方法。
以20~30 ng DNA为模板, 使用包含“CCTACGRRBGCASCAGKVRVGAA”序列的上游引物和包含“GGACTACNVGGGTWTCTAATCC”[13]序列的下游引物扩增V3和V4区。采用25 µL反应体系(Trans Start Buffer 2.5 µL, dNTPs 2 µL, Primers 1 µL×2, TransStart Taq DNA 0.5 µL, 模板DNA 20 ng, ddH2O补至总体系25 µL), 反应条件为: 94℃, 3 min; 94℃, 5 s, 57℃, 90 s, 72℃, 10 s, 24个循环; 72℃, 5 min进行扩增。PCR产物用1.5%琼脂糖凝胶电泳检测。另外, 通过PCR向16S rDNA的PCR产物末端加上带有Index接头, 以便进行测序。
通过Qubit3.0 Fluorometer (Invitrogen, CarIsbad, CA)检测文库浓度。将文库定量到10 nmol/L, 按Illumina MiSeq (Illumina, San Diego, CA, USA)仪器使用说明书进行PE250双端测序, 由MiSeq自带Miseq Control Software (MCS)读取序列。测序委托苏州金唯智生物科技有限公司进行, 对原始序列进行预处理选取高质量的序列进行数据分析。
1.4 数据分析MiSeq测序结果用QIIME (1.9.1)进行质量筛选。使用UCHIME识别和移除嵌合体序列[14]。使用VSEARCH(1.9.6)进行序列聚类(序列相似性设为97%), 比对的参考数据库是Silva_132 16S rRNA database(http://www.arb-sliva.de)[15]。根据归一化的OTU丰度计算Shannon指数和Chao 1指数等α多样性指数[16]。在归一化基础上, 基于Bray-Curtis差异系数研究β多样性[17]。
在R(3.5.2)软件包(https://www.r-project.org)中进行了统计分析和绘图。使用vegan(2.5.2)进行NMDS排序, 应用ANOSIM (analysis of similarities)相似性分析对不同群落间的相似性进行检验, 使用vegan(2.5.2)和ggplot2进行冗余分析(RDA)确定细菌群落与环境因子之间的关系。利用SPSS(19.0)统计软件(IBM Corporation, USA)进行Pearson相关分析, 确定环境因素与浮游细菌多样性(Shannon指数与Simpson指数)以及优势细菌丰度之间的关系。
2 结果与分析 2.1 浮游细菌的群落组成珠江河网所有样品的覆盖度(goods-coverage)为99.7%(表 1), 表明细菌测序深度已达到较高水平, 数据可靠。所有样品的有效序列数在39072~ 76392之间, OTU数目在359~658之间(序列同源性为97%)(表 1), 所有样本均达到饱和阶段, 表明样本足以进行后续数据分析(图 2)。
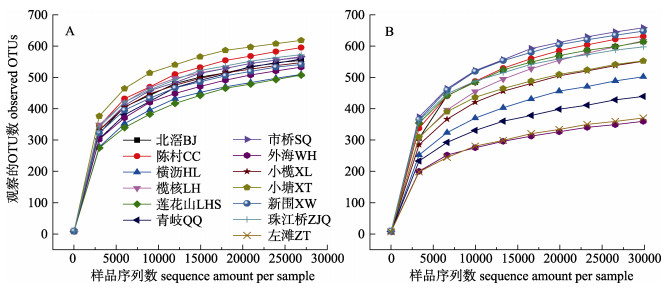
|
图 2 珠江河网冬季浮游细菌稀释曲线 A. 2017年11月; B. 2018年1月. Fig.2 Rarefaction curve of planktonic bacteria in winter in Pearl River Delta A. November 2017; B. January 2018. |
|
|
表 1 珠江河网调查区域冬季浮游细菌序列特征 Tab.1 Sequence characteristics of planktonic bacteria in winter in the survey area of the Pearl River Delta |
在珠江河网冬季的两次采样中, 除未分类的细菌外, 2017年11月的样品序列主要分布于23个门类, 2018年1月样品主要分布于29个门类, 13个门类为两次样品所共有。其中变形菌门(Proteobacteria)、放线菌门(Actinobacteria)、拟杆菌门(Bacteroidetes)、蓝细菌门(Cyanobacteria)是冬季相对丰度较高的门类, 不同时间采集样品物种的相对丰度有所改变。其中变形菌门和拟杆菌门的相对丰度在2017年11月(分别为42.63%~ 68.68%, 5.03%~14.81%)均低于2018年1月(分别为48.9%~88.58%, 3.07%~40.19%); 放线菌的相对丰度在2017年11月(7.2%~43.98%)高于2018年1月(1.93%~22.85%); 蓝细菌门在2018年1月的左滩(ZT_Jan, 0.25%)、外海(WH_Jan, 0.13%)及青岐(QQ_Jan, 0.26%)观察到较低的丰度, 在其余站点样品中的丰度均大于1%(图 3)。
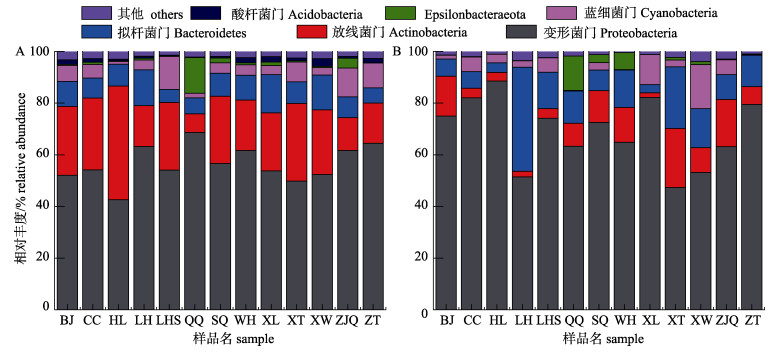
|
图 3 门分类水平的珠江河网冬季浮游细菌相对丰度
A. 2017年11月; B. 2018年1月. BJ:北滘, CC:陈村, HL:横沥, LH:榄核, LHS:莲花山, QQ:青岐, SQ:市桥, WH:外海, XL:小榄, XT:小塘, XW:新围, ZJQ:珠江桥, ZT:左滩. Fig.3 Relative abundance of planktonic bacteria in Pearl River Delta at phylum level A. November 2017; B. January 2018. BJ: Beijiao, CC: Chencun, HL: Hengli, LH: Lanhe, LHS: Lianhuashan, QQ: Qingqi, SQ: Shiqiao, WH: Waihai, XL: Xiaolan, XT: Xiaotang, XW: Xinwei, ZJQ: Zhujiangqiao, ZT: Zuotan. |
统计两次样品在纲分类水平上相对丰度最高的30个物种, 发现24个物种为两次样品所共有。其中γ-变形菌纲(γ-Proteobacteria)、放线菌纲(Actinobacteria)、α-变形菌纲(α-Proteobacteria)、拟杆菌纲(Bacteroidia)、酸微菌纲(Acidimicrobiia)和产氧光细菌纲(Oxyphotobacteria)是珠江河网的优势种(图 4)。
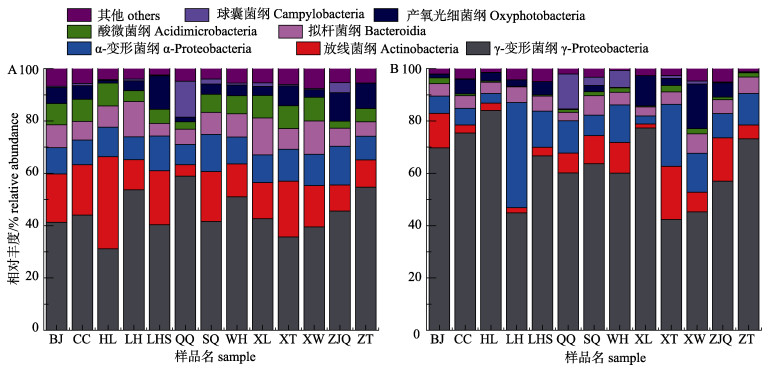
|
图 4 纲分类水平的珠江河网冬季浮游细菌相对丰度
A. 2017年11月; B. 2018年1月. BJ:北滘, CC:陈村, HL:横沥, LH:榄核, LHS:莲花山, QQ:青岐, SQ:市桥, WH:外海, XL:小榄, XT:小塘, XW:新围, ZJQ:珠江桥, ZT:左滩. Fig.4 Relative abundance of planktonic bacteria in Pearl River Delta at class level A. November 2017; B. January 2018. BJ: Beijiao, CC: Chencun, HL: Hengli, LH: Lanhe, LHS: Lianhuashan, QQ: Qingqi, SQ: Shiqiao, WH: Waihai, XL: Xiaolan, XT: Xiaotang, XW: Xinwei, ZJQ: Zhujiangqiao, ZT: Zuotan. |
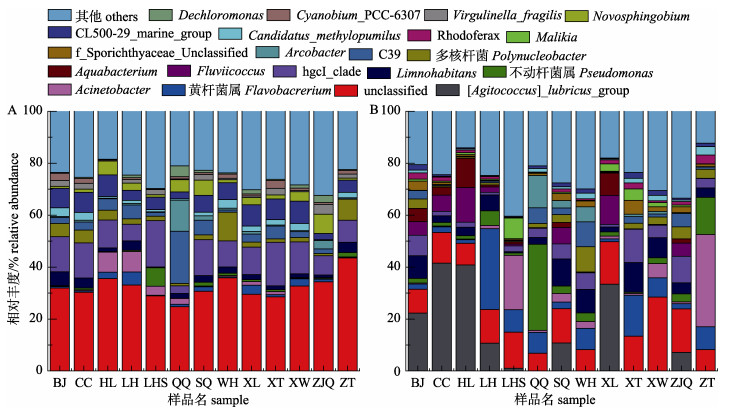
2017年11月样品在属分类水平上有15个属的相对丰度大于1%, Unclassified占据总量的32.29%, hgcI_clade (Actinobacteria, 11.11%)、C39 (Proteobacteria, 4.29%)、Flavobacterium (Bacteroidetes, 1.55%)和Virgulinella_frgilis (Cyanobacteria, 1.15%)分别是放线菌门、变形菌门、拟杆菌门和蓝细菌门相对丰度最高的属。2018年1月样品在属分类水平上有16个属的相对丰度大于1%, [Agitococcus]_lubricus_group (Proteobacteria, 12.90%)、Flavobacterium (Bacteroidetes, 7.62%)和hgcI_clade (Actinobacteria, 4.84%)分别是变形菌门、拟杆菌门以及放线菌门相对丰度最高的属(图 5)。
|
图 5 属分类水平的珠江河网冬季浮游细菌相对丰度
A. 2017年11月; B. 2018年1月. BJ:北滘, CC:陈村, HL:横沥, LH:榄核, LHS:莲花山, QQ:青岐, SQ:市桥, WH:外海, XL:小榄, XT:小塘, XW:新围, ZJQ:珠江桥, ZT:左滩. Fig.5 Relative abundance of planktonic bacteria in Pearl River Delta at genus level A. November 2017; B. January 2018. BJ: Beijiao, CC: Chencun, HL: Hengli, LH: Lanhe, LHS: Lianhuashan, QQ: Qingqi, SQ: Shiqiao, WH: Waihai, XL: Xiaolan, XT: Xiaotang, XW: Xinwei, ZJQ: Zhujiangqiao, ZT: Zuotan. |
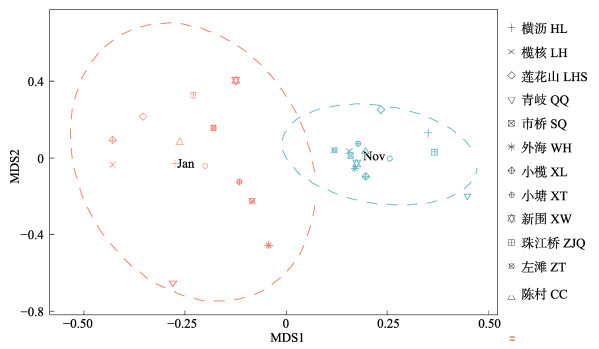
对冬季珠江河网浮游细菌群落进行多样性分析, 结果表明两次采样的ACE指数分别为622.69±26.07、638.04±93.16, Shannon指数分别为6.78±0.29、6.23±0.71(表 2), 表明珠江河网冬季的细菌丰富度和多样性均处于较高水平, 且群落的稳定性较高。基于Bray-Curtis差异系数做NMDS分析(图 6), 结果显示珠江河网的浮游细菌群落结构在不同的采样时期存在显著差异,而采样地点之间的细菌群落不存在差异。利用ANOSIM分析对NMDS分析结果进行检验, 将样品按照时间进行分组,组间存在较大差异(R=0.88, P=0.001), 按照地点进行分组, 组内差异显著大于组间差异(R=-0.21,P=1)。进一步表明了珠江河网的浮游细菌群落存在时间上的显著差异, 而不存在地点上的显著差异。
|
图 6 珠江河网冬季浮游细菌群落NMDS分析 Fig.6 NMDS analysis of planktonic bacteria community in winter in Pearl River delta |
|
|
表 2 珠江河网冬季浮游细菌α多样性指数 Tab.2 α-diversity indices of planktonic bacteria in winter in Pearl River Delta |
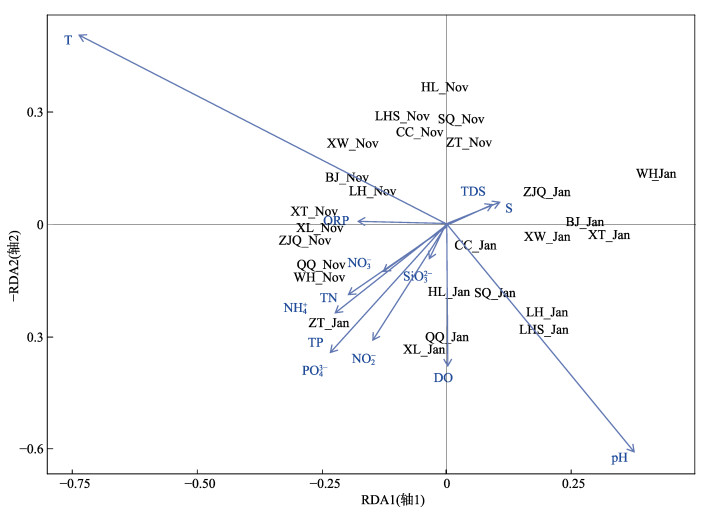
珠江河网水环境因子检测结果见表 3。利用RDA和Pearson分析环境因子与细菌群落之间的关系。在RDA分析中, RDA 1和RDA 2解释67.15%的变异(图 7)。分析结果表明温度(r2=0.577, P < 0.01)和pH(r2=0.513, P < 0.01)是影响浮游细菌群落的主要环境因子。此外溶解氧(r2=0.181, P > 0.05), ORP(r2=0.144, P > 0.05), 总磷(r2=0.023, P > 0.05), 总氮(r2=0.047, P > 0.05), 磷酸盐(r2=0.04, P > 0.05), 硝酸盐(r2=0.012, P > 0.05), 亚硝酸盐(r2=0.006, P > 0.05), 铵盐(r2=0.019, P > 0.05), 硅酸盐(r2=0.083, P > 0.05)与浮游细菌群落之间没有表现出显著相关性。
|
图 7 珠江河网冬季浮游细菌群落RDA分析
Nov: 2017年11月, Jan: 2018年1月. BJ:北滘, CC:陈村, HL:横沥, LH:榄核, LHS:莲花山, QQ:青岐, SQ:市桥, WH:外海, XL:小榄, XT:小塘, XW:新围, ZJQ:珠江桥, ZT:左滩.
Fig.7 RDA analysis of planktonic bacteria community in winter in the Pearl River Delta
Nov: November 2017; Jan: January 2018. BJ: Beijiao, CC: Chencun, HL: Hengli, LH: Lanhe, LHS: Lianhuashan, QQ: Qingqi, SQ: Shiqiao, WH: Waihai, XL: Xiaolan, XT: Xiaotang, XW: Xinwei, ZJQ: Zhujiangqiao, ZT: Zuotan. |
|
|
表 3 珠江河网水体环境因子检测结果 Tab.3 Monitoring results of water environmental factors in the Pearl River Delta |
Pearson相关分析结果如表 4所示, 其中温度与γ-变形菌纲(R=0.43, P < 0.05)、放线菌纲(R=0.436, P < 0.05)以及α-变形菌纲(R=0.777, P < 0.01)的丰度显著正相关,pH与放线菌纲(R=-0.469, P < 0.05)、α-变形菌(R=‒0.76, P < 0.01)的丰度以及多样性指数(Shannon: R=‒0.504, P < 0.01; Simpson: R=‒0.515, P < 0.01)显著负相关, 溶解氧与α-变形菌纲的丰度显著负相关(R=‒0.467, P < 0.05)。其他环境因子与优势种的丰度以及群落多样性不存在显著相关性(P > 0.05)。
|
|
表 4 珠江河网细菌群落多样性和优势种丰度与环境特征的相关性分析 Tab.4 Correlation analysis between bacteria community diversity, abundance of dominant species and environmental characteristics in the Pearl River Delta |
变形菌门、拟杆菌门、放线菌门和蓝细菌门是珠江河网的优势类群, 与淡水生态系统的典型类群一致[18-19]。在Zwart等[19]的研究中, 以不同的河流和湖泊作为研究对象, 利用16S rDNA分析淡水生态系统中的浮游细菌, 发现变形菌门、蓝细菌门、放线菌门和疣微菌门以及拟杆菌门中的黄杆菌纲是典型的淡水细菌类群, 并且认为河流和湖泊的浮游细菌群落不同于土壤、沉积物等邻近环境中的细菌群落, 并且这些浮游细菌分布在世界各地不同的淡水生态系统中。
珠江河网的细菌群落组成以及各类群的相对丰度与洞庭湖细菌群落模式极为相似[20], 其中变形菌门是优势类群, γ-变形菌纲是优势亚群。值得注意的是, γ-变形菌在城区河段[21]、污水尾水[22]以及养殖水体[23]等营养水平较高的水体中占优势, 珠江河网γ-变形菌在细菌群落中的分布情况与上述研究极为相似。通过对水环境监测结果显示珠江河网的总磷处于Ⅳ~Ⅴ类水平, 总氮处于劣Ⅴ类水平, 进一步表明了珠江河网的水体环境的营养水平较高(GB3838-2002)。
hgcI_clade是珠江河网放线菌门的优势属, 具有较强的吸收碳、氮化合物的能力, 对低氧具有耐受性, 能利用水中的低浓度溶解有机碳[24-25], 对水体的净化有重要作用。黄杆菌属是拟杆菌门的优势属, 黄杆菌属具有广泛的溶藻能力, 对水质的提高有重要作用[26]。C39、[Agit ococcus]_ lubricus_group是变形菌门的优势属, 但是对其在水域生态系统中作用研究相对较少。此外, 蓝细菌门是珠江河网相对丰度稍低的优势群, 在属分类水平上不占显著优势, 原因可能是冬季的光照强度以及温度较低, 影响了蓝细菌的生长的繁殖。
3.2 珠江河网浮游细菌群落差异及其影响因素本研究发现珠江河网的浮游细菌群落在同一季节的不同时期存在显著性差异, Lv等[27]对黄河三角洲的研究发现菌群的差异出现在冬季与其他季节之间, Li等[28]发现珠江口的细菌群落在丰水期和枯水期出现显著差异。细菌群落发生的改变可能与环境因子的波动存在着一定关系。
利用RDA分析可直接表示环境因子与细菌群落的相关情况。本研究对珠江河网环境因子与细菌群落的分析表明, 温度和pH是影响浮游细菌群落的主要环境因子, 这个结果与Lindstr m等[29]对湖泊研究的结论一致。
在相关的研究中, 细菌群落的组成变化更多地是由温度驱动[30-31]。温度是影响浮游细菌群落结构及丰度季节变化的重要参数[32]。在本研究中, γ-变形菌、放线菌和α-变形菌的丰度均与温度呈显著正相关关系, 表明在冬季(调查期间温度15~26℃), 随着温度的升高, γ-变形菌、放线菌与α-变形菌的丰度都有显著增加的趋势。在适宜的温度范围内, 随着温度升高细菌的生产力大大增加, 不同细菌类群的最适生长温度不同, 处于最适温度的细菌生产力是其他温度的2~11倍[30]。低温作为一种环境压力在冬季影响着微生物的群落结构及活性[33]。在本研究中, 拟杆菌在温度较低环境下的相对丰度要高于温度稍高的环境, 这与在黄河三角洲观察到的情况相似[22], 表明拟杆菌对低温的耐受性较强, 在低温环境中具有较高的竞争能力[34]。
在本研究中, pH是影响浮游细菌群落结构及多样性的另一个主要环境因子, 其对细菌的多样性及群落结构的显著影响在多种生态系统中均有体现[35-37]。在中性或碱性培养基中接种湖泊细菌的实验研究发现, 酸性湖泊的细菌在培养基中的生长受到抑制[38], 表明了细菌对不同pH的偏好。在本研究水域, pH处于7.56~8.55, 放线菌纲、α-变形菌的丰度随着pH的增加而呈显著降低, 细菌群落多样性也是如此。但是对pH在细菌群落中的代谢影响机制还未进行更加深入的研究。
微生物的生长除了温度、pH等影响因素外, 营养因素N、P至关重要, 许多相关研究也发现营养因素对浮游细菌群落结构有很大影响。例如, 北海湖研究表明, 当TN含量为0.83~1.67 mg/L, TP含量为0.04~0.11 mg/L时, 营养盐浓度增加, 能显著增加蓝细菌的丰度[26]; 对钱塘江的研究发现, OTUs随着硝酸盐浓度的增加而显著降低(r=-0.805, P < 0.005), 铵盐(r=-0.764, P < 0.005)和总无机氮(r=-0.826, P < 0.005)与Shannon指数呈显著负相关[35]; 对太湖的研究发现, 在TN含量为2.97~9.67 mg/L, TP含量为0.08~0.11 mg/L时, 细菌数量随水体营养水平上升而增加, 群落多样性则随水体营养水平的增加呈现显著下降的趋势[39]。但在本研究中, 浮游细菌群落结构与N、P不存在显著相关性。对比发现, 珠江河网的TN含量为2.28~3.43 mg/L, TP含量为0.14~0.38 mg/L, 高于北海湖的TN、TP含量水平以及太湖的TP水平。在营养水平较低的水体中, 无机营养物质的含量可能是细菌生长的限制因子, 就珠江河网而言, N、P水平较高, 已不是细菌生长的限制因子, 因此对细菌的群落结构的影响未达到显著水平。
4 结论运用16S rDNA扩增子技术研究珠江河网浮游细菌群落结构, 结果表明变形菌门、放线菌门、拟杆菌门和蓝细菌门是珠江河网冬季丰度较高的门类, 优势种群依次为γ-变形菌、放线菌和α-变形菌。珠江河网细菌群落具有较高的多样性, 在研究区域内细菌群落组成较为稳定。随着采样时间的不同, 细菌群落结构出现显著性差异。其中温度和pH是影响珠江河网冬季浮游细菌群落的主要环境因子,溶解氧与α-变形菌纲的丰度显著负相关; 珠江河网的N、P水平较高, 已不是细菌生长的限制因子, 因此对细菌的群落结构的影响未达到显著水平。
| [1] |
Findlay S. Stream microbial ecology[J]. Journal of the North American Benthological Society, 2010, 29(1): 170-181. DOI:10.1899/09-023.1 |
| [2] |
Giuliano L, De Domenico M, De Domenico E, et al. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing[J]. Microbial Ecology, 1999, 37(2): 77-85. DOI:10.1007/s002489900132 |
| [3] |
Wang J, Peng J F, Song Y H, et al. Seasonal changes of microbial community distribution in sediments of Hun River[J]. Research of Environmental Science, 2016, 29(2): 202-210. [王佳, 彭剑峰, 宋永会, 等. 浑河底泥微生物群落的季节性变化特征[J]. 环境科学研究, 2016, 29(2): 202-210.] |
| [4] |
Bowen J L, Morrison H G, Hobbie J E, et al. Salt marsh sediment diversity:a test of the variability of the rare biosphere among environmental replicates[J]. The ISME Journal, 2012, 6(11): 2014-2023. DOI:10.1038/ismej.2012.47 |
| [5] |
Guo L D. Progress of microbial species diversity research in China[J]. Biodiversity Science, 2012, 20(5): 572-580. [郭良栋. 中国微生物物种多样性研究进展[J]. 生物多样性, 2012, 20(5): 572-580.] |
| [6] |
Lu L, Jia Z J. Urease gene-containing archaea dominate autotrophic ammonia oxidation in two acid soils[J]. Environmental Microbiology, 2013, 15(6): 1795-1809. DOI:10.1111/emi.2013.15.issue-6 |
| [7] |
Kim S J, Park S J, Cha I T, et al. Metabolic versatility of toluene-degrding, iron-reducing bacteria in tidal flat sediment, characterized by stable isotope probing-based metagenomic analysis[J]. Environmental Microbiology, 2014, 16(1): 189-204. DOI:10.1111/emi.2014.16.issue-1 |
| [8] |
Yu L Y, Zhang W J, Liu L M, et al. Determining microeukaryotic plankton community around Xiamen Island, Southeast China, using Illumina MiSeq and PCR-DGGE techniques[J]. PLoS ONE, 2015, 10(5): e0127721. DOI:10.1371/journal.pone.0127721 |
| [9] |
Wu X, Xi W Y, Ye W J, et al. Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences[J]. FEMS Microbiology Ecology, 2010, 61(1): 85-96. |
| [10] |
Gao Y, Lai Z N, Zeng Y Y, et al. Community structure of copepods and the relationship with aquatic environmental factors in the Pearl River Delta[J]. Journal of Fishery Sciences of China, 2015, 22(2): 302-310. [高原, 赖子尼, 曾艳艺, 等. 珠江三角洲河网桡足类群落结构及其与水环境因子的关系[J]. 中国水产科学, 2015, 22(2): 302-310.] |
| [11] |
Huang X D. Discussion on ecological status of water and countermeasures of ecological restoration of small and middle-sized rivers in the Pearl River Delta of Guangdong Province[J]. Guangdong Water Resources and Hydropower, 2016(5): 16-19. [黄显东. 广东省珠江三角洲地区中小河流水生态现状及修复对策初探[J]. 广东水利水电, 2016(5): 16-19.] |
| [12] |
Wang C, Li X H, Lai Z N, et al. Temporal and spatial pattern of the phytoplankton biomass in the Pearl River Delta[J]. Acta Ecologica Sinica, 2013, 33(18): 5835-5847. [王超, 李新辉, 赖子尼, 等. 珠三角河网浮游植物生物量的时空特征[J]. 生态学报, 2013, 33(18): 5833-5847.] |
| [13] |
Walters W, Hyde E R, Berg-Lyons D, et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys[J]. mSystems, 2016, 1(1): e00009-15. |
| [14] |
Edgar R C, Haas B J, Clemente J C, et al. UCHIME improves sensitivity and speed of chimera detection[J]. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [15] |
Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project:improved data processing and web-based tools[J]. Nucleic Acids Research, 2013, 41: D590-D596. |
| [16] |
Wang Y, Zhang R, He Z L, et al. Functional gene diversity and metabolic potential of the microbial community in an estuary-shelf environment[J]. Frontiers in Microbiology, 2017, 8: Article No. 1153.
|
| [17] |
Lundin D, Severin I, Logue J B, et al. Which sequencing depth is sufficient to describe patterns in bacterial α-and β-diversity?[J]. Environmental Microbiology Reports, 2012, 4(3): 367-372. DOI:10.1111/emi4.2012.4.issue-3 |
| [18] |
Wu X, Xi W Y, Ye W J, et al. Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences[J]. FEMS Microbiology Ecology, 2007, 61(1): 85-96. DOI:10.1111/fem.2007.61.issue-1 |
| [19] |
Zwart G, Crump B C, Agterveld M, et al. Typical freshwater bacteria:an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers[J]. Aquatic Microbial Ecology, 2002, 28(2): 141-155. |
| [20] |
Huang W, Jiang X. Profiling of sediment microbial community in Dongting Lake before and after impoundment of the Three Gorges Dam[J]. International Journal of Environmental Research and Public Health, 2016, 13(6): Article No. 617.
|
| [21] |
Tang J, Xu X R, Shang C Y, et al. Association of bacterial diversty in city area of Nanming river with environmental factors[J]. Acta Microbiologica Sinica, 2015, 55(8): 1050-1059. [唐婧, 徐小蓉, 商传禹, 等. 南明河城区河段细菌多样性与环境因子的关系[J]. 微生物学报, 2015, 55(8): 1050-1059.] |
| [22] |
Xu A L, Ren J, Song Z W, et al. Microbial community of municipal discharges in a sewage treatment plant[J]. Environmental Science, 2014, 35(9): 3473-3479. [徐爱玲, 任杰, 宋志文, 等. 污水处理厂尾水细菌群落结构分析[J]. 环境科学, 2014, 35(9): 3473-3479.] |
| [23] |
Sun Z L, Xuan Y M, Zhang H, et al. Bacterial diversity in the Penaeus vannamei Boone intestine and aquaculture environment[J]. Journal of Fishery Sciences of China, 2016, 23(3): 594-605. [孙振丽, 宣引明, 张浩, 等. 南美白对虾养殖环境及其肠道细菌多样性分析[J]. 中国水产科学, 2016, 23(3): 594-605.] |
| [24] |
Ghylin T W, Garcia S L, Moya F, et al. Comparative single-cell genomics reveals potential ecological niches for the freshwater acI Actinobacteria lineage[J]. The ISME Journal, 2014, 8(12): 2503-2516. DOI:10.1038/ismej.2014.135 |
| [25] |
Liu J W, Fu B B, Yang H M, et al. Phylogenetic shifts of bacterioplankton community composition along the Pearl Estuary: the potential impact of hypoxia and nutrients[J]. Frontiers in Microbiology, 2015, 6: Article No. 64.
|
| [26] |
Zhang Y J, Li K, Zhu H R, et al. Community structure of microorganisms and its seasonal variation in Beihai Lake[J]. Environmental Science, 2017, 38(8): 3319-3329. [张雅洁, 李珂, 朱浩然, 等. 北海胡微生物群落结构随季节变化特征[J]. 环境科学, 2017, 38(8): 3319-3329.] |
| [27] |
Lv X F, Ma B, Yu J B, et al. Bacterial community structure and function shift along a successional series of tidal flats in the Yellow River Delta[J]. Scientific Reports, 2016, 6: Article No. 36550.
|
| [28] |
Li J Y, Jiang X, Jing Z Y, et al. Spatial and seasonal distributions of bacterioplankton in the Pearl River Estuary:The combined effects of riverine inputs, temperature, and phytoplankton[J]. Marine Pollution Bulletin, 2017, 125: 199-207. DOI:10.1016/j.marpolbul.2017.08.026 |
| [29] |
Lindstr m E S, Kamst-Van Agterveld M P, Zwart G. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time[J]. Applied and Environmental Microbiology, 2005, 71(12): 8201-8206. DOI:10.1128/AEM.71.12.8201-8206.2005 |
| [30] |
Adams H E, Crump B C, Kling G W. Temperature controls on aquatic bacterial production and community dynamics in arctic lakes and streams[J]. Environmental Microbiology, 2010, 12(5): 1319-1333. DOI:10.1111/emi.2010.12.issue-5 |
| [31] |
Sun Z, Li G, Wang C W, et al. Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir[J]. Scientific Reports, 2014, 4: Article No. 6966.
|
| [32] |
Zhang M L, Yu N, Chen L Q, et al. Structure and seasonal dynamics of bacterial communities in three urban rivers in China[J]. Aquatic Sciences, 2012, 74(1): 113-120. DOI:10.1007/s00027-011-0201-z |
| [33] |
Han G X, Yu J B, Li H B, et al. Winter soil respiration from different vegetation patches in the Yellow River Delta[J]. Environmental Management, 2012, 50: 39-49. DOI:10.1007/s00267-012-9869-7 |
| [34] |
Georges A A, El-Swais H, Craig S E, et al. Metaproteomic analysis of a winter to spring succession in coastal northwest Atlantic Ocean microbial plankton[J]. The ISME Journal, 2014, 8(6): 1303-1313. |
| [35] |
Liu S, Ren H X, Shen L D, et al. pH levels drive bacterial community structure in sediments of the Qiantang River as determined by 454 pyrosequencing[J]. Frontiers in Microbiology, 2015, 6: Article No. 285.
|
| [36] |
Hollister E B, Engledow A S, Hammett A J, et al. Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments[J]. The ISME Journal, 2010, 4: 829-838. DOI:10.1038/ismej.2010.3 |
| [37] |
Shen C C, Xiong J B, Zhang H Y, et al. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain[J]. Soil Biology and Biochemistry, 2013, 57: 204-211. DOI:10.1016/j.soilbio.2012.07.013 |
| [38] |
Langenheder S, Lindstr m E S, Tranvik L J. Weak coupling between community composition and functioning of aquatic bacteria[J]. Limnology and Oceanography, 2005, 50(3): 957-967. DOI:10.4319/lo.2005.50.3.0957 |
| [39] |
Feng S, Qin B Q, Gao G. Response of bacterial communities to eutrophic water in Lake Taihu[J]. Acta Scientiae Circumstantiae, 2002, 27(11): 1823-1829. [冯胜, 秦伯强, 高光. 细菌群落结构对书体富营养化的响应[J]. 环境科学学报, 2002, 27(11): 1823-1829.] |
 2019, Vol. 26
2019, Vol. 26

