2. 中国水产科学研究院南海水产研究所, 热带水产研究开发中心, 农业农村部南海渔业资源开发利用重点实验室, 广东 广州 510300;
3. 仲恺农业工程学院, 健康养殖创新研究院, 广东 广州 510225;
4. 珠海市现代农业发展中心, 广东 珠海 519000
2. Tropical Fisheries Research and Development Center, South China Sea Fisheries Research Institute, Chinese Acad-emy of Fishery Sciences; Key Laboratory of South China Sea Fishery Resources Development and Utilization, Min-istry of Agriculture and Rural Affairs, Guangzhou 510300, China;
3. Innovative Institute of Animal Healthy Breeding, Zhongkai University of Agriculture and Engineering, Guangzhou 510225, China;
4. Modern Agricultural Development Center of Zhuhai City, Zhuhai 519000, China
花鲈(Lateolabrax maculatus), 又称海鲈, 是中国第二大海水养殖鱼类, 在广东珠海养殖面积超过16 km2, 产量分别约占广东省的80%、全国的50%[1], 是广东省特色水产养殖鱼类之一。随着养殖集约化程度的增加, 病害问题限制了花鲈产业的可持续发展。其中, 杀鱼爱德华菌(Edwardsiella piscicidas)是花鲈养殖的主要病原之一, 此菌在世界各地养殖区域爆发流行[2-3], 广泛存在于环境和宿主中, 可感染多种经济鱼类[4-6]。
杀鱼爱德华菌与迟缓爱德华菌(E. tarda)有许多相同的表型特征[7], 目前仍存在一些菌株被误归为迟缓爱德华菌的现象。Griffin等[8-9]通过gyrB来对爱德华菌进行菌株分类, 将迟缓爱德华菌和杀鱼爱德华菌进行区分。自1995年Yamamoto等[10]首次用gyrB基因鉴别恶臭假单胞菌(Pseudomonas putida)近缘种后, 越来越多的学者用该基因进行菌种鉴定。gyrB基因作为在分类学中被应用最广泛的管家基因, 其遗传密码子具有优良的兼并性, 在DNA序列发生替换时可保证氨基酸序列稳定, 其表达水平明显高于16S rDNA基因, 可更正确地区分近缘种。此外, gyrB基因不易在水平间转移, 可作为系统发育分析中的靶基因[11]。
细菌耐药性的出现和迅速传播使其成为公共卫生安全的焦点[12], 病原菌的耐药机制已经成为临床微生物学中重要且广泛的研究内容[13]。许多研究发现, 病原菌抗生素耐药性机制的主要决定因素是从其他生物水平基因转移获得的, 同时在对病原菌另一大致病因素——毒力研究中同样发现水平基因转移的共同基本特征。毒力和抗生素抗性都可被认为是细菌探索和开拓生存新环境的方法, 这些方法使细菌的基因组表现出高度可塑性, 所以抗生素抗性和毒力在机制研究中同时被称为“量子跃迁的进化”[14]。有研究表明抗生素抗性基因和毒力基因可以处于同个可移动元件(如质粒、转座子、噬菌体、整合子和基因簇)中[15-16], 如迟缓爱德华菌中发现pCK41质粒同时存在耐药基因和毒力基因[17]; 大肠杆菌中发现pCERC3质粒同时具有毒力和耐药基因[18]; 多重耐药(MDR)外排泵能外排抗生素和群体感应信号等物质[19]。进一步研究表明, 毒力和抗生素之间存在相关性联系[20], Ghorbel等[21]研究发现毒力模式与抗菌药物图谱存在显著相关性; Azzam等[22]研究表明MAR与污水生态系统细菌毒力具有强相关性; Vila等[23]发现细菌获得抗生素耐药时可导致毒力下降, 对其耐药机制研究表明大部分菌株通过改变自身表达或蛋白结构来增加耐药性[24]。目前对于其相关性的研究大多集中在人类医学和畜牧行业中, 在水产养殖领域研究较少, 所以通过对杀鱼爱德华菌毒力与耐药的相关性研究, 可以为该菌的抗生素使用提供指导。
本研究团队基于gyrB基因序列信息, 对分离自2018年4—12月广东省珠海市池塘养殖花鲈的杀鱼爱德华菌菌株耐药谱和毒力特征评价, 为阐明花鲈源杀鱼爱德华菌的流行趋势和耐药规律奠定基础, 并对耐药性和毒力相关性研究提供数据支持。
1 材料与方法 1.1 菌株分离及鉴定采样时间为2018年4―12月, 样品采自广东省珠海市养殖患病的花鲈内脏器官, 具体细菌分离方法参照鱼病调查手册[25], 分离菌株进行革兰氏染色, 通过16S rDNA通用引物和gyrB基因引物进行分子鉴定(表 1), 对鉴定为杀鱼爱德华菌的菌株进行后续实验。
|
|
表 1 基因引物序列 Tab.1 Gene primer sequence |
将鉴定获得的87株杀鱼爱德华菌制备菌悬液, 菌液浓度为1×108 CFU/mL, 采用纸片扩散法(K-B法)进行药敏试验, 记录药敏纸片抑菌圈的直径。药敏纸片为16种常见的抗生素(均购于杭州微生物试剂有限公司):阿莫西林(amoxicillin, AMO, 20 μg)、麦迪霉素(midecamycin, MD, 30 μg)、利福平(rifampin, RFP, 5 μg)、青霉素(penicillin, P, 10 μg)、复方新诺明(norepinephrine, SXT, 1.25 μg)、诺氟沙星(norfloxacin, NOR, 10 μg)、红霉素(erythromycin, E, 15 μg)、氟苯尼考(florfenicol, FFC, 30 μg)、氯霉素(chloramphenicol, C, 30 μg)、呋喃唑酮(furazolidone, FUR, 100 μg)、新霉素(neomycin, N, 10 μg)、庆大霉素(gentamicin, GM, 10 μg)、磺胺异恶唑(sulfamethoxazole, FS, 300 μg)、土霉素(oxytetracycline, OT, 30 μg)、多西环素(doxycycline, DO, 30 μg)、恩诺沙星(enrofloxacin, ENR, 30 μg)。参照美国临床和实验室标准协会抗生素敏感试验标准分析菌株耐药谱(表 2)[26]。将杀鱼爱德华菌对每一种抗生素的耐药情况以S、I、R记录, 则每株受试菌株对所有抗生素形成唯一的耐药谱, 对所有耐药谱进行归类并命名。计算耐药率及多重抗生素耐药指数(MARI), MARI=a/bc其中a代表某来源细菌的总抗菌药物耐药值, b代表检测的抗菌药物种类数目, c代表某来源细菌菌株[27], 分析比较耐药谱型所包括的抗生素及菌株量差异, 用Origin软件进行作图分析。
|
|
表 2 药物敏感实验判定标准 Tab.2 Criteria for antibiotics sensitivity test |
选择体长4 cm左右且健康的AB品系斑马鱼, (25±1) ℃恒温暂养一周。将培养好的87株杀鱼爱德华菌用生理盐水稀释至浓度约为3×105 CFU/mL, 腹腔注射剂量20 μl/尾, 每株菌注射30尾鱼, 分3个平行组, 记录注射后2周内的死亡率。
1.4 数据处理将杀鱼爱德华菌对16种抗生素的耐药情况进行赋值(R=4, I=2, S=0), 对每种抗生素耐药情况(R=4, I=2, S=0)与斑马鱼死亡率结果结合, 利用IBM SPSS 22进行Spearman相关性分析, 得到其相关性结果。
2 结果与分析 2.1 细菌分离鉴定对优势菌进行革兰氏染色, 发现有96株菌株在显微镜下呈现红色短杆状。通过16S rDNA通用引物和gyrB基因引物进行扩增, 测序后分析基因序列的同源性, 发现杀鱼爱德华菌占87株, 迟缓爱德华菌仅占9株。大部分采样鱼呈现鱼眼部肿大、眼球缺失、头骨处皮肤溃烂、腹水以及内脏器官肿大等症状。
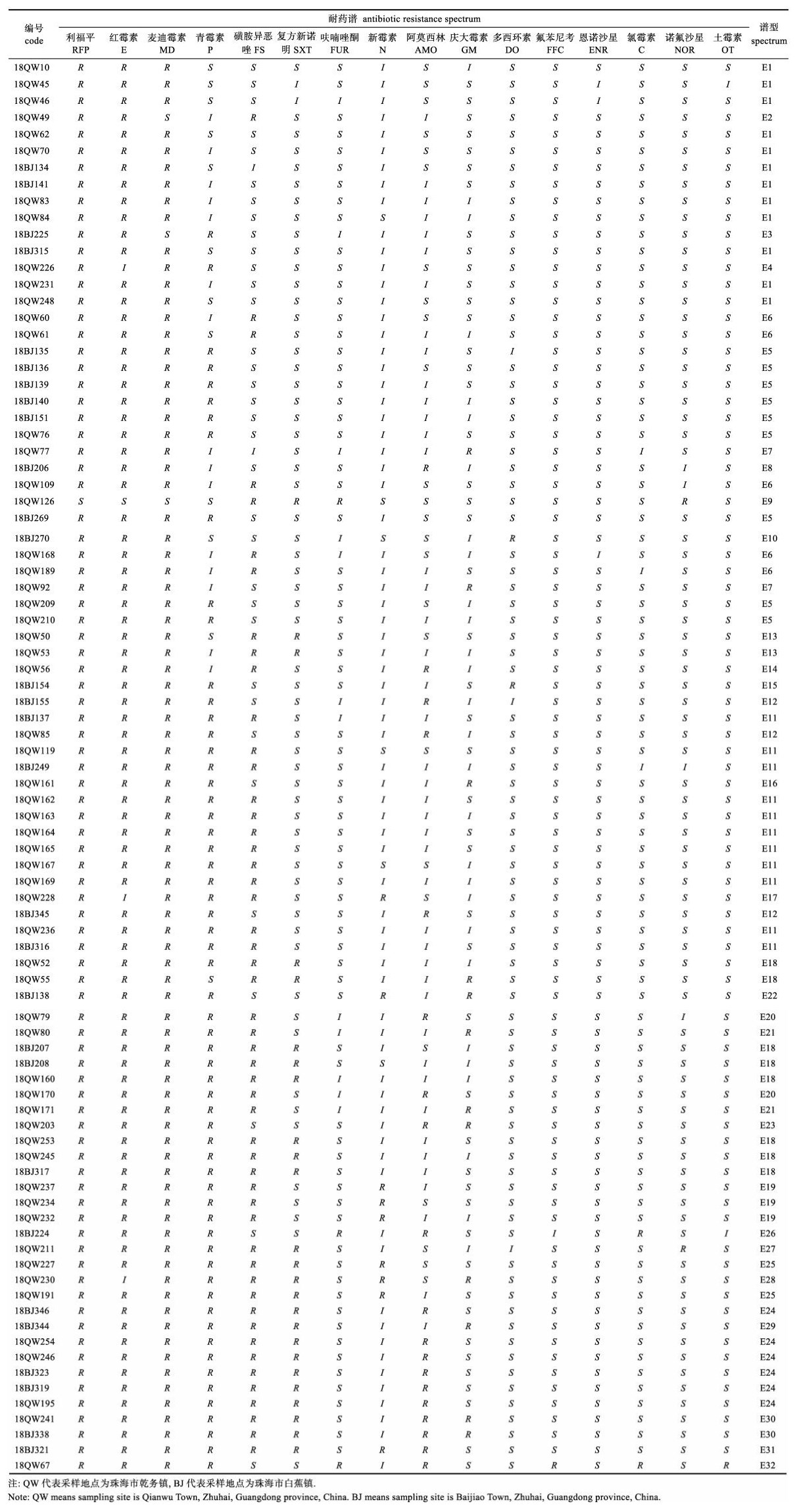
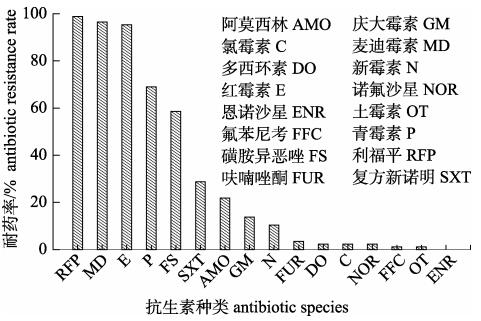
2.2 杀鱼爱德华菌耐药谱87株杀鱼爱德华菌对16种抗生素的耐药存在差异(图 1), 其中对麦迪霉素、红霉素和利福平的耐药率超过了90%及以上, 对青霉素、磺胺异恶唑耐药率在80%~50%, 对庆大霉素、阿莫西林和复方新诺明耐药率均低于50%~10%, 对多西环素、氟苯尼考、恩诺沙星、氯霉素、诺氟沙星和土霉素的耐药率低于10%。杀鱼爱德华菌的耐药谱型共32种, 分为E1~E32 (表 3), 谱型丰富度为36.78%。E1~E3型谱型对3种抗生素耐药, 包含14菌株, 占总菌数的16.09%; E4~E10谱型对4种抗生素耐药, 包含20菌株, 占总菌数的22.98%; E11~E17谱型对5种抗生素耐药, 包含20菌株, 占总菌数的22.98%; E18~E23谱型对6种抗生素耐药, 包含17菌株, 占总菌数的19.10%; E24~ E29谱型对7种抗生素耐药, 包含12菌株, 占总菌数的13.80%; E30、E31谱型对8种抗生素耐药, 包含3菌株, 占总菌数的3.45%; E32对9种抗生素耐药, 包含1菌株, 占总菌数的1.60%。87株菌MARI指数为0.423, 抗4~6种抗生素的菌株居多, 占到总菌数的64.36%。
|
图 1 87株杀鱼爱德华菌菌株对16种抗生素的耐药性 Fig.1 Antibiotic resistance of 87 Edwardsiella piscicida strains to 16 antibiotics |
|
|
表 3 87株杀鱼爱德华菌的耐药谱型 Tab.3 Antibiotic resistance spectrum of 87 strains of Edwardsiella piscicida |
杀鱼爱德华菌中对斑马鱼致死率大于90%有20株菌, 占总杀鱼爱德华菌株数的22.99%, 对斑马鱼致死率在80%~90%的有17株菌, 占19.54%, 对斑马鱼致死率小于30%有11株菌, 占12.64% (表 4)。
|
|
表 4 87株杀鱼爱德华菌对斑马鱼致死率 Tab.4 Mortality of Danio rerioto 87 Edwardsiella piscicida strains |
通过Spearman相关性分析, 可知斑马鱼死亡率与庆大霉素耐药性呈正相关(P < 0.05), 相关系数0.255, 与恩诺沙星、氟苯尼考(P < 0.05)和土霉素(P < 0.01)耐药性呈负相关, 相关系数分别为0.237、0.245、0.297 (表 5)。
|
|
表 5 杀鱼爱德华菌毒力与耐药相关性分析 Tab.5 Analysis of the relationship between virulence and antibiotics resistance of Edwardsiella piscicida |
爱德华菌属起初分为迟缓爱德华菌, 鲶爱德华菌(E. ictaluri)和保科爱德华菌(E. hoshinae) 3个种, 后有学者发现迟缓爱德华菌又可再分为迟缓爱德华菌、杀鱼爱德华菌和鳗鲡爱德华菌(E. anguillarum)[28-29]。近些年来, 杀鱼爱德华菌因从迟缓爱德华菌中分出而成为一种新的鱼类病原菌, 可引起鱼体的败血症[30]、表皮腐烂瘀斑、深部皮肤溃疡暴露、骨头坏死[4]以及多个器官出现肉芽肿病变[5-6]等病症。造成上述症状感染和产生致病性主要是由毒力基因和毒力调节系统造成的[31], 包括溶血素[32]、软骨素酶、三型分泌系统[33]、六型分泌系统等。而本研究分离出杀鱼爱德华菌的花鲈鱼体不完全呈现上述典型的病理症状, 可能是该病原所致的不同临床症状与宿主或菌株间毒力基因(系统)存在差异有关。
在水产养殖中, 细菌对抗生素的耐药性主要来自抗生素的选择性压力[34], 药物的频繁和过量使用, 会导致细菌出现自适应突变, 即出现多重耐药甚至“超级细菌”[35]。以前研究发现, 酰胺醇类抗生素[36]、四环素类[37-38]和喹诺酮类[39-40]抗生素在中国海域检出率较高。而本实验结果显示, 杀鱼爱德华菌对多西环素、氟苯尼考、恩诺沙星、氯霉素、诺氟沙星和土霉素的耐药性较低。显然从鱼体中分离的菌株部分耐药性与水体的抗生素检测结果不同, 可能与近年来相关部门提倡科学用药和禁止养殖中使用氯霉素、诺氟沙星和土霉素等抗生素在降低药物耐药率方面发挥一定作用, 在杀鱼爱德华菌上得到了体现。本研究中耐药谱显示杀鱼爱德华菌对红霉素、麦迪霉素和利福平有较高的耐药性。而Xie等[41]和Xu等[42]报道红霉素和麦迪霉素在珠三角水域和养殖区域的检出率较高, 同样Ma等[43]发现环境中普遍存在利福平耐药, 即从鱼体中分离的菌株耐药性与水体的抗生素检测结果相同, 与之前结果矛盾, 推测与杀鱼爱德华菌对大环内酯类和利福霉素类抗生素耐药性的持续保持有关, 有待后续进一步研究。
多重抗生素耐药(MARI)是分析指定菌群耐药性的极佳工具, 当菌株暴露于常用抗菌药物, MARI > 0.2, 而少用或不用抗菌药物时, MARI < 0.2[27]。多重抗生素耐药结果显示, 杀鱼爱德华菌MARI指数为0.423, 说明该菌在珠海养殖区处于抗生素高危暴露状态, 由于本实验暂未对其耐药机制进行研究, 故珠海花鲈源杀鱼爱德华菌高MARI指数形成原因暂不明确。
3.2 杀鱼爱德华菌毒力与耐药相关性分析斑马鱼(Danio rerio)是一种研究病原体感染和宿主免疫反应的模式动物[44], 已建立了斑马鱼的气单胞菌[45]、链球菌[46]、沙门氏菌[47]和爱德华菌感染模型[48]。本研究揭示, 分离的87株杀鱼爱德华菌菌株对斑马鱼毒力存在一定的差异, 证实杀鱼爱德华菌含有不同毒力类型的菌株[49]。细菌毒力因子主要存在于染色体基因簇中, 或存在于质粒和噬菌体等遗传辅助元件中, 其扩散同细菌耐药基因的扩散存在相关的机制。有学者发现毒力与耐药基因在一定程度上可以相互传播和转化[50-51]。并且在编码细菌素[52]、铁载体[53]、细胞毒素[54]和黏附因子[55]的基因中都存在抗生素抗性。研究发现ESPfm (肠球菌表面蛋白)与氨苄耐药呈正相关[56]; 细菌生物膜与耐药性呈正相关[57]; 细胞膜蛋白失活导致毒力与耐药均下降[58]。所以研究表明细菌抗生素耐药和毒力存在遗传连锁和表达共轭。这与本实验发现的杀鱼爱德华菌毒力与庆大霉素抗性呈正相关(P < 0.05)的结果相似。当然, 也存在相反的表型, 如耐青霉素的肺炎链球菌菌株致病性可能低于敏感菌株[59]; 多重耐药导致毒力下降[60]; 对利福平耐药导致毒力下降[61]; 从氟喹诺酮耐药菌株中发现毒力基因表达下降[62-63]。对此, 许多学者提出了“生物适应性成本”的概念, 即抗性突变引起细菌的适合度下降[64], 所以当可移动元件接合到细菌中, 可能发生异常的细菌基因调控即生物适应性成本增加。当病原菌抗生素耐药性增加时, 由于新的遗传决定因子可能会使病原菌产生生物适应性成本, 这可能会导致细菌适应度下降, 从而使细菌毒性降低。所以, 当病原菌抗生素耐药和毒力之间由于生物适应性成本的增加出现负相关情况, 并且大多数情况表明主要的相关性为负相关[65-67], 这与本研究中杀鱼爱德华菌毒力与恩诺沙星、氟苯尼考(P < 0.05)和土霉素抗性(P < 0.01)呈负相关的结果一致。但长远来看, 细菌由于获得性耐药导致调控异常(毒力减弱)会慢慢通过细菌的补偿突变进行回复, 甚至在某些个别的情况中会出现不会改变细菌适应度的突变[68-69]。虽然目前在研究中分析大部分抗生素与毒力呈现负相关, 但由于细菌的补偿突变, 终究会形成高毒力, 高耐药的菌株[70-72]。本研究第一次在杀鱼爱德华菌上提出毒力与耐药之间的相关性概念, 并发现杀鱼爱德华菌耐药与毒力之间大多呈现负相关, 对杀鱼爱德华菌耐药和毒力相关性的分子机制尚有待进一步深入研究。
| [1] |
Wen H S, Zhang M Z, Li J F, et al. Research progress of aquaculture industry and its seed engineering in spotted sea bass (Lateolabrax maculatus) of China[J]. Fishery Information & Strategy, 2016, 31(2): 105-111. [温海深, 张美昭, 李吉方, 等. 我国花鲈养殖产业现状与种子工程研究进展[J]. 渔业信息与战略, 2016, 31(2): 105-111.] |
| [2] |
Camus A, Dill J, McDermott A, et al. Edwardsiella piscicida-associated septicaemia in a blotched fantail stingray Taeniura meyeni (Müeller & Henle)[J]. Journal of Fish Diseases, 2016, 39(9): 1125-1131. DOI:10.1111/jfd.12435 |
| [3] |
Shafiei S, Viljamaa-Dirks S, Sundell K, et al. Recovery of Edwardsiella piscicida from farmed whitefish, Coregonus lavaretus (L.), in Finland[J]. Aquaculture, 2016, 654: 19-26. |
| [4] |
Reichley S R, Waldbieser G C, Tekedar H C, et al. Complete genome sequence of Edwardsiella piscicida isolate S11-285 recovered from channel catfish (Ictalurus punctatus) in Mississippi, USA[J]. Genome Announcements, 2016, 4(6): e01259-16. |
| [5] |
Oguro K, Tamura K, Yamane J, et al. Draft genome sequences of two genetic variant strains of Edwardsiella piscicida, JF1305 and RSB1309, isolated from olive flounder (Paralichythys olivaceus) and Red Sea Bream (Pagrus major) cultured in Japan, respectively[J]. Genome Announcements, 2014, 2(3): e00546-14. |
| [6] |
Fogelson S B, Petty B D, Reichley S R, et al. Histologic and molecular characterization of Edwardsiella piscicida infection in largemouth bass (Micropterus salmoides)[J]. Journal of Veterinary Diagnostic Investigation, 2016, 28(3): 338-344. DOI:10.1177/1040638716637639 |
| [7] |
Buján N, Mohammed H, Balboa S, et al. Genetic studies to re-affiliate Edwardsiella tarda fish isolates to Edwardsiella piscicida and Edwardsiella anguillarum species[J]. Systematic and Applied Microbiology, 2018, 41(1): 30-37. DOI:10.1016/j.syapm.2017.09.004 |
| [8] |
Griffin M J, Ware C, Quiniou S M, et al. Edwardsiella piscicida identified in the southeastern USA by gyrB sequence, species-specific and repetitive sequence-mediated PCR[J]. Diseases of Aquatic Organisms, 2014, 108(1): 23-35. DOI:10.3354/dao02687 |
| [9] |
Griffin M J, Petty B D, Ware C, et al. Recovery and confirmation of Edwardsiella piscicida from a black crappie Pomoxis nigromaculatus (Lesueur, 1829)[J]. Journal of Fish Diseases, 2019, 42(10): 1457-1461. DOI:10.1111/jfd.13056 |
| [10] |
Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains[J]. Applied and Environmental Microbiology, 1995, 61(10): 3768. DOI:10.1128/AEM.61.10.3768-3768.1995 |
| [11] |
Soler L. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes[J]. International Journal of Systematic and Evolutionary Microbiology, 2004, 54(5): 1511-1519. DOI:10.1099/ijs.0.03048-0 |
| [12] |
Wu J J, Su Y L, Deng Y Q, et al. Spatial and temporal variation of antibiotic resistance in marine fish cage-culture area of Guangdong, China[J]. Environmental Pollution, 2019, 246: 463-471. DOI:10.1016/j.envpol.2018.12.024 |
| [13] |
Martínez J L, Baquero F. Interactions among strategies associated with bacterial infection:Pathogenicity, epidemicity, and antibiotic resistance[J]. Clinical Microbiology Reviews, 2002, 15(4): 647-679. DOI:10.1128/CMR.15.4.647-679.2002 |
| [14] |
Groisman E A, Ochman H. Pathogenicity islands:Bacterial evolution in quantum leaps[J]. Cell, 1996, 87(5): 791-794. DOI:10.1016/S0092-8674(00)81985-6 |
| [15] |
Reid C J, McKinnon J, Djordjevic S P. Clonal ST131-H22Escherichia coli strains from a healthy pig and a human urinary tract infection carry highly similar resistance and virulence plasmids[J]. Microbial Genomics, 2019, 5(9): 000295. |
| [16] |
Villa L, Carattoli A. Integrons and transposons on the Salmonella enterica serovar typhimurium virulence plasmid[J]. Antimicrobial Agents and Chemotherapy, 2005, 49(3): 1194-1197. DOI:10.1128/AAC.49.3.1194-1197.2005 |
| [17] |
Yu J E, Cho M Y, Kim J W, et al. Large antibiotic-resistance plasmid of Edwardsiella tarda contributes to virulence in fish[J]. Microbial Pathogenesis, 2012, 52(5): 259-266. DOI:10.1016/j.micpath.2012.01.006 |
| [18] |
Moran R A, Holt K E, Hall R M. pCERC3 from a commensal ST95Escherichia coli:A ColV virulence-multiresistance plasmid carrying a sul3-associated class 1 integron[J]. Plasmid, 2016, 84-85: 11-19. DOI:10.1016/j.plasmid.2016.02.002 |
| [19] |
Zgurskaya H I, Nikaido H. Multidrug resistance mechanisms:Drug efflux across two membranes[J]. Molecular Microbiology, 2000, 37(2): 219-225. DOI:10.1046/j.1365-2958.2000.01926.x |
| [20] |
Rathnayake I U, Hargreaves M, Huygens F. Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates[J]. Systematic and Applied Microbiology, 2012, 35(5): 326-333. DOI:10.1016/j.syapm.2012.05.004 |
| [21] |
Ghorbel D, Hadrich I, Neji S, et al. Detection of virulence factors and antifungal susceptibility of human and avian Aspergillus flavus isolates[J]. Journal De Mycologie Médicale, 2019, 29(4): 292-302. DOI:10.1016/j.mycmed.2019.100900 |
| [22] |
Azzam M I, Ezzat S M, Othman B A, et al. Antibiotics resistance phenomenon and virulence ability in bacteria from water environment[J]. Water Science, 2017, 31(2): 109-121. DOI:10.1016/j.wsj.2017.10.001 |
| [23] |
Vila J, Simon K, Ruiz J, et al. Are quinolone-resistant uropathogenic Escherichia coli less virulent?[J]. The Journal of Infectious Diseases, 2002, 186(7): 1039-1042. DOI:10.1086/342955 |
| [24] |
Wang B B, Li B, Liang Y, et al. Pleiotropic effects of temperature-regulated 2-OH-lauroytransferase (PA0011) on Pseudomonas aeruginosa antibiotic resistance, virulence and type III secretion system[J]. Microbial Pathogenesis, 2016, 91: 5-17. DOI:10.1016/j.micpath.2015.11.003 |
| [25] |
Fish Disease Research Office of Hubei Institute of Hydrobiology. Fish Disease Survey Manual[M]. Second Edition. Shanghai: Shanghai Scientific & Technical Publishers, 1961. [湖北省水生生物研究所鱼病研究室. 鱼病调查手册[M]. 第2版. 上海: 上海科学技术出版社, 1961.]
|
| [26] |
CLSI. Performance standards for antimicrobial disk susceptibility Testing, approved standard, M100-S22[S]. 11th Edition, CLSI document M02-A11. Wayne: Clinical and Laboratory Standards Institute, 2012: 1-53.
|
| [27] |
Titilawo Y, Sibanda T, Obi L, et al. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water[J]. Environmental Science and Pollution Research, 2015, 22(14): 10969-10980. DOI:10.1007/s11356-014-3887-3 |
| [28] |
Abayneh T, Colquhoun D J, Sø rum H. Multi-locus sequence analysis (MLSA) of Edwardsiella tarda isolates from fish[J]. Veterinary Microbiology, 2012, 158(3-4): 367-375. DOI:10.1016/j.vetmic.2012.03.006 |
| [29] |
Griffin M J, Quiniou S M, Cody T, et al. Comparative analysis of Edwardsiella isolates from fish in the eastern United States identifies two distinct genetic taxa amongst organisms phenotypically classified as E. tarda[J]. Veterinary Microbiology, 2013, 165(3-4): 358-372. DOI:10.1016/j.vetmic.2013.03.027 |
| [30] |
Esteve C, Alcaide E. Seasonal recovery of Edwardsiella piscicida from wild European eels and natural waters:Isolation methods, virulence and reservoirs[J]. Journal of Fish Diseases, 2018, 41(11): 1613-1623. DOI:10.1111/jfd.12867 |
| [31] |
Liu Y, Oshima S I, Kurohara K, et al. Vaccine efficacy of recombinant GAPDH of Edwardsiella tarda against edwardsiellosis[J]. Microbiology and Immunology, 2005, 49(7): 605-612. DOI:10.1111/j.1348-0421.2005.tb03652.x |
| [32] |
Hirono I, Tange N, Aoki T. Iron-regulated haemolysin gene from Edwardsiella tarda[J]. Molecular Microbiology, 1997, 24(4): 851-856. DOI:10.1046/j.1365-2958.1997.3971760.x |
| [33] |
Tan Y P, Zheng J, Tung S L, et al. Role of type III secretion in Edwardsiella tarda virulence[J]. Microbiology, 2005, 151(7): 2301-2313. DOI:10.1099/mic.0.28005-0 |
| [34] |
Carvalho M J, Martínez-Murcia A, Esteves A C, et al. Phylogenetic diversity, antibiotic resistance and virulence traits of Aeromonas spp. from untreated waters for human consumption[J]. International Journal of Food Microbiology, 2012, 159(3): 230-239. |
| [35] |
Huo T I. The first case of multidrug-resistant NDM-1-harboring Enterobacteriaceae in Taiwan:Here comes the superbacteria![J]. Journal of the Chinese Medical Association, 2010, 73(11): 557-558. DOI:10.1016/S1726-4901(10)70121-0 |
| [36] |
Yoo M H, Huh M D, Kim E H, et al. Characterization of chloramphenicol acetyltransferase gene by multiplex polymerase chain reaction in multidrug-resistant strains isolated from aquatic environments[J]. Aquaculture, 2003, 217(1-4): 11-21. DOI:10.1016/S0044-8486(02)00169-2 |
| [37] |
Dang H Y, Zhao J Y, Song L S, et al. Molecular characterizations of chloramphenicol-and oxytetracycline-resistant bacteria and resistance genes in mariculture waters of China[J]. Marine Pollution Bulletin, 2009, 58(7): 987-994. DOI:10.1016/j.marpolbul.2009.02.016 |
| [38] |
Tamminen M, Karkman A, Lõhmus A, et al. Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure[J]. Environmental Science & Technology, 2011, 45(2): 386-391. |
| [39] |
Nie X P, He X T, Yang Y T, et al. Investigation of quinolones in aquaculture environment of the Pearl River Delta[J]. Environmental Science, 2009, 30(1): 266-270. [聂湘平, 何秀婷, 杨永涛, 等. 珠江三角洲养殖水体中喹诺酮类药物残留分析[J]. 环境科学, 2009, 30(1): 266-270. DOI:10.3321/j.issn:0250-3301.2009.01.045] |
| [40] |
He X T, Deng M C, Wang Q, et al. Residues and health risk assessment of quinolones and sulfonamides in cultured fish from Pearl River Delta, China[J]. Aquaculture, 2016, 458: 38-46. DOI:10.1016/j.aquaculture.2016.02.006 |
| [41] |
Xie H W, Hao H S, Xu N, et al. Pharmaceuticals and personal care products in water, sediments, aquatic organisms, and fish feeds in the Pearl River Delta:Occurrence, distribution, potential sources, and health risk assessment[J]. Science of the Total Environment, 2019, 659: 230-239. DOI:10.1016/j.scitotenv.2018.12.222 |
| [42] |
Xu W H, Yan W, Li X D, et al. Antibiotics in riverine runoff of the Pearl River Delta and Pearl River Estuary, China:concentrations, mass loading and ecological risks[J]. Environmental Pollution, 2013, 182: 402-407. DOI:10.1016/j.envpol.2013.08.004 |
| [43] |
Ma L P, Li B, Zhang T. Abundant rifampin resistance genes and significant correlations of antibiotic resistance genes and plasmids in various environments revealed by metagenomic analysis[J]. Applied Microbiology and Biotechnology, 2014, 98(11): 5195-5204. DOI:10.1007/s00253-014-5511-3 |
| [44] |
Lin C, Lin C N, Wang Y C, et al. The role of TGF-β signaling and apoptosis in innate and adaptive immunity in zebrafish:A systems biology approach[J]. BMC Systems Biology, 2014, 8: 116. DOI:10.1186/s12918-014-0116-0 |
| [45] |
Chandrarathna H P S U, Nikapitiya C, Dananjaya S H S, et al. Outcome of co-infection with opportunistic and multidrug resistant Aeromonas hydrophila and A. veronii in zebrafish:identification, characterization, pathogenicity and immune responses[J]. . veronii in zebrafish:identification, characterization, pathogenicity and immune responses, 2018, 80: 573-581. |
| [46] |
Patterson H, Saralahti A, Parikka M, et al. Adult zebrafish model of bacterial meningitis in Streptococcus agalactiae infection[J]. Developmental & Comparative Immunology, 2012, 38(3): 447-455. |
| [47] |
Li Y Y, Wang T, Gao S, et al. Salmonella plasmid virulence gene spvB enhances bacterial virulence by inhibiting autophagy in a zebrafish infection model[J]. Fish & Shellfish Immunology, 2016, 49: 252-259. |
| [48] |
Liu X H, Chang X Y, Wu H Z, et al. Role of intestinal inflammation in predisposition of Edwardsiella tarda infection in zebrafish (Danio rerio)[J]. Fish & Shellfish Immunology, 2014, 41(2): 271-278. |
| [49] |
Yin K Y, Guan Y P, Ma R Q, et al. Critical role for a promoter discriminator in RpoS control of virulence in Edwardsiella piscicida[J]. PLoS Pathogens, 2018, 14(8): e1007272. DOI:10.1371/journal.ppat.1007272 |
| [50] |
Alexander T W, Yanke J L, Reuter T, et al. Longitudinal characterization of antimicrobial resistance genes in feces shed from cattle fed different subtherapeutic antibiotics[J]. BMC Microbiology, 2011, 11(1): 19. |
| [51] |
Barton M D. Antibiotic use in animal feed and its impact on human health[J]. Nutrition Research Reviews, 2000, 13(2): 279-299. |
| [52] |
Chelliah R, Wei S, Park B J, et al. New perspectives on mega plasmid sequence (poh1) in Bacillus thuringiensis ATCC 10792 harbouring antimicrobial, insecticidal and antibiotic resistance genes[J]. Microbial Pathogenesis, 2019, 126: 14-18. DOI:10.1016/j.micpath.2018.10.013 |
| [53] |
Zhang W L, Zhang Y, Wang X X, et al. Siderophores in clinical isolates of Klebsiella pneumoniae promote ciprofloxacin resistance by inhibiting the oxidative stress[J]. Biochemical and Biophysical Research Communications, 2017, 491(3): 855-861. DOI:10.1016/j.bbrc.2017.04.108 |
| [54] |
Carlson S A, Meyerholz D K, Stabel T J, et al. Secretion of a putative cytotoxin in multiple antibiotic resistant Salmonella enterica serotype typhimurium phagetype DT104[J]. Microbial Pathogenesis, 2001, 31(4): 201-204. DOI:10.1006/mpat.2001.0461 |
| [55] |
Laporta M Z, Silva M L, Scaletsky I C, et al. Plasmids coding for drug resistance and localized adherence to HeLa cells in enteropathogenic Escherichia coli O55:H- and O55:H6[J]. Infection and Immunity, 1986, 51(2): 715-717. DOI:10.1128/IAI.51.2.715-717.1986 |
| [56] |
Billströ m H, Lund B, Sullivan Ả, et al. Virulence and antimicrobial resistance in clinical Enterococcus faecium[J]. International Journal of Antimicrobial Agents, 2008, 32(5): 374-377. DOI:10.1016/j.ijantimicag.2008.04.026 |
| [57] |
Pereira M F, Rossi C C, Seide L E, et al. Antimicrobial resistance, biofilm formation and virulence reveal Actinobacillus pleuropneumoniae strains' pathogenicity complexity[J]. Research in Veterinary Science, 2018, 118: 498-501. DOI:10.1016/j.rvsc.2018.05.003 |
| [58] |
Liang Y, Guo Z S, Gao L, et al. The role of the temperature-regulated acyltransferase (PA3242) on growth, antibiotic resistance and virulence in Pseudomonas aeruginosa[J]. Microbial Pathogenesis, 2016, 101: 126-135. DOI:10.1016/j.micpath.2016.09.019 |
| [59] |
Azoulay-Dupuis E, Rieux V, Muffat-Joly M, et al. Relationship between capsular type, penicillin susceptibility, and virulence of human Streptococcus pneumoniae isolates in mice[J]. Antimicrobial Agents and Chemotherapy, 2000, 44(6): 1575-1577. DOI:10.1128/AAC.44.6.1575-1577.2000 |
| [60] |
Lehtolainen T, Shwimmer A, Shpigel N Y, et al. In vitro antimicrobial susceptibility of Escherichia coli isolates from clinical bovine mastitis in Finland and Israel[J]. Journal of Dairy Science, 2003, 86(12): 3927-3932. DOI:10.3168/jds.S0022-0302(03)74001-6 |
| [61] |
O'Neill A J, Huovinen T, Fishwick C W G, et al. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence[J]. Antimicrobial Agents and Chemotherapy, 2006, 50(1): 298-309. DOI:10.1128/AAC.50.1.298-309.2006 |
| [62] |
Schaeffer A J. Decreased invasive capacity of quinolone-resistant Escherichia coli in patients with urinary tract infections[J]. Journal of Urology, 2002, 168(1): 393-394. |
| [63] |
Liu M C, Wu C M, Liu Y C, et al. Identification, susceptibility, and detection of integron-gene cassettes of Arcanobacterium pyogenes in bovine endometritis[J]. Journal of Dairy Science, 2009, 92(8): 3659-3666. DOI:10.3168/jds.2008-1756 |
| [64] |
Andersson D I. The biological cost of mutational antibiotic resistance:Any practical conclusions?[J]. Current Opinion in Microbiology, 2006, 9(5): 461-465. DOI:10.1016/j.mib.2006.07.002 |
| [65] |
Toutounji M, Tokajian S, Khalaf R A. Genotypic and phenotypic characterization of Candida albicans Lebanese hospital isolates resistant and sensitive to caspofungin[J]. Fungal Genetics and Biology, 2019, 127: 12-22. DOI:10.1016/j.fgb.2019.02.008 |
| [66] |
Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates[J]. Clinical Microbiology and Infection, 2010, 16(12): 1770-1775. DOI:10.1111/j.1469-0691.2010.03177.x |
| [67] |
Qin X H, Hu F P, Wu S, et al. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains[J]. PLoS ONE, 2013, 8(4): e61169. DOI:10.1371/journal.pone.0061169 |
| [68] |
Criswell D, Tobiason V L, Lodmell J S, et al. Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi[J]. Antimicrobial Agents and Chemotherapy, 2006, 50(2): 445-452. DOI:10.1128/AAC.50.2.445-452.2006 |
| [69] |
Foucault M L, Depardieu F, Courvalin P, et al. Inducible expression eliminates the fitness cost of vancomycin resistance in Enterococci[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(39): 16964-16969. DOI:10.1073/pnas.1006855107 |
| [70] |
McCallum N, Berger-Bä chi B, Senn M M. Regulation of antibiotic resistance in Staphylococcus aureus[J]. International Journal of Medical Microbiology, 2010, 300(2-3): 118-129. DOI:10.1016/j.ijmm.2009.08.015 |
| [71] |
Saddler C A, Wu Y, Valckenborgh F, et al. Epidemiological control of drug resistance and compensatory mutation under resistance testing and second-line therapy[J]. Epidemics, 2013, 5(4): 164-173. DOI:10.1016/j.epidem.2013.08.002 |
| [72] |
Durã o P, Balbontín R, Gordo I. Evolutionary mechanisms shaping the maintenance of antibiotic resistance[J]. Trends in Microbiology, 2018, 26(8): 677-691. DOI:10.1016/j.tim.2018.01.005 |
 2020, Vol. 27
2020, Vol. 27